Some evolutionary genetic models predict that small populations are under continuous threat from accumulating deleterious mutations in their genomes, while other models suggest small populations can evolve to be robust to these effects. To date, there is almost no empirical evidence that describes the impact of long-term population size on genome evolution.
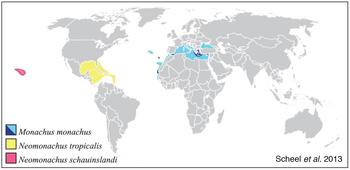
In order to explore this fundamental evolutionary relationship, Stephen Gaughran, a graduate student in the lab, is generating whole genome sequencing data for multiple species of seals with very different demographic histories. The Hawaiian monk seal (Neomonachus schauinslandi) and the Mediterranean monk seal (Monachus monachus) are both endangered species, and Stephen’s demographic modelling work suggests that these species have existed in relatively small populations for much of their evolutionary history. Other seal species, for example the Weddell seal (Leptonychotes weddellii), live in much larger populations. Finally, some species such as the Northern elephant seal (Mirounga angustirostris) and the Caribbean monk seal (Neomonachus tropicalis) have experienced recent extreme bottlenecks, leading to extinction in the case of the Caribbean monk seal.
Stephen’s comparative genomics work models how these very different demographic histories have affected the molecular evolution of these species, including the rate of mutation accumulation and the patterns of amino acid substitution in protein-coding genes. This work will have important implications for the future of conservation genetics, as researchers and managers begin to use whole genome data of endangered species to inform conservation decisions.